By Kate Freund, University of Minnesota
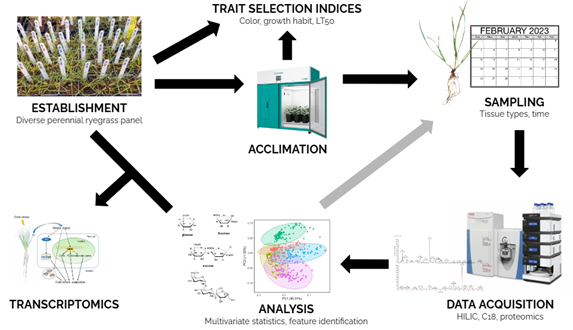
As discussed in part 1 of this blog, to begin exploring the metabolome of perennial ryegrass in relation to cold tolerance (Figure 1), data must first be collected. There are a variety of analytical platforms on which this can be done, and all are based on the separation and detection of metabolites. Common separation techniques include gas chromatography (GC) and liquid chromatography (LC). Detection and characterization methods include mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy. For our work on cold tolerance in perennial ryegrass we will be utilizing high performance liquid chromatography-mass spectrometry (HPLC-MS; Figure 2). HPLC-MS is a combined system in which liquid chromatography first separates compounds and then mass spectrometry measures the mass/charge (m/z) ratio to determine masses of the molecules.

After samples are extracted, they are injected into the HPLC system and forced by the mobile phase (typically a buffered solvent) through a column packed with a stationary phase (porous solid material with a specialized coating) specific to the type of separation that is desired. The interface between the HPLC column and the MS is an electrospray ion source that forces the separated sample through a needle while applying a high voltage, causing the molecules to become charged ions. These ions are then filtered through the MS and their masses are analyzed. Multivariate statistical analyses such as principal component analysis (PCA) and partial least squares regression (PLS) are then used to model relationships between the metabolites and the samples. From these analyses, biomarkers (measurable indicators for a specific phenotype) for cold tolerance can be determined, and further work can be conducted to identify the compounds and their functions. The use of HPLC-MS is suitable for the detection and analysis of a wide range of metabolites, making it a favored analytical technique for untargeted metabolomics studies on perennial ryegrass.
Our current work is an extension and expansion of earlier work conducted in the Turfgrass Science program at the University of Minnesota. Previous studies have used metabolomics approaches to select plants for enhanced rust resistance (Koeritz 2014), to explore the effects of fungicide application on hard fescue in correlation with transcriptomic analyses, and to screen perennial ryegrass accessions for improved freezing tolerance. The next stage for this research is to use metabolomics, along with other omics techniques, to guide the selection of perennial ryegrass germplasm from a larger array of accessions and gain a deeper understanding of cold tolerance.

To develop perennial ryegrass cultivars with improved freezing tolerance, we must explore what factors contribute to superior freezing tolerance. Past research on perennial ryegrass and other grasses has identified specific genes which when expressed result in the production of cryoprotective proteins, amino acids, soluble sugars, fructans, phenylpropanoids, and phytohormones that are associated with increased membrane stability, slowed growth, and reduced freezing. We are currently curating a large diverse panel of perennial ryegrass accessions from around the world (Figure 3) with the goals of 1) developing standardized methods to assess appropriate time points and tissues to sample for metabolomics, and 2) associating freezing tolerance with various metabolites including sugars, proteins, amino acids, and other specialized metabolites. Additionally, we will be providing plant material for transcriptomic and proteomic analyses in tandem with our metabolomic analyses to identify and correlate candidate genes associated with cold tolerance. Overall, this work will guide the development of cold-tolerant germplasm while also establishing standardized procedures for expanding this work to other species and desirable traits.
References
Koeritz, E. (2014). Improving Sustainability of Perennial Ryegrass (Lolium perenne) Seed Production: Integrated crop management practices and the development of a novel metabolomics-assisted technique to select for resistance to rust (Puccinia) pathogens. Retrieved from the University of Minnesota Digital Conservancy, https://hdl.handle.net/11299/173928
Miao, C., Zhang, Y., Bai, X., and Qin, T. (2022). Insights into the Response of Perennial Ryegrass to Abiotic Stress: Underlying Survival Strategies and Adaptation Mechanisms. Life, 12(6), 860.